 |
GURLS++
2.0.00
C++ Implementation of GURLS Matlab Toolbox
|
 |
GURLS++
2.0.00
C++ Implementation of GURLS Matlab Toolbox
|
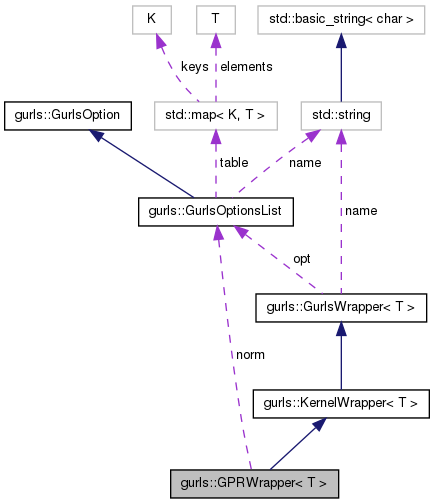
GPRWrapper is the sub-class of GurlsWrapper that implements ...
#include <gprwrapper.h>
Public Types | |
| enum | KernelType { RBF, LINEAR } |
| enum | ProblemType { CLASSIFICATION, REGRESSION } |
Public Member Functions | |
| gMat2D< T > * | eval (const gMat2D< T > &X) |
| Estimates label for an input matrix. | |
| gMat2D< T > * | eval (const gMat2D< T > &X, gMat2D< T > &vars) |
| Estimates label for an input matrix. | |
| virtual T | eval (const gVec< T > &X, unsigned long *index=NULL) |
| Estimates label for a new input point. | |
| virtual const GurlsOptionsList & | getOpt () const |
| Returns a const reference to the options structure. | |
| GPRWrapper (const std::string &name) | |
| Constructor. | |
| virtual void | loadModel (const std::string &fileName) |
| Loads a computed model from a file. | |
| virtual void | saveModel (const std::string &fileName) |
| Saves the computed model to file. | |
| virtual void | setKernelType (KernelType value) |
| virtual void | setNparams (unsigned long value) |
| virtual void | setNSigma (unsigned long value) |
| virtual void | setParam (double value) |
| virtual void | setProblemType (ProblemType value) |
| virtual void | setSigma (double value) |
| virtual void | setSplitProportion (double value) |
| void | train (const gMat2D< T > &X, const gMat2D< T > &y) |
| Initial parameter selection and training. | |
| ~GPRWrapper () | |
| Destructor. | |
Protected Member Functions | |
| virtual bool | trainedModel () |
| Checks if model has already been trained. | |
Protected Attributes | |
| KernelType | kType |
| Kernel type used in train and eval. | |
| std::string | name |
| Name of the options structure. | |
| GurlsOptionsList * | norm |
| GurlsOptionsList * | opt |
| Options structure where information about initial training is stored. | |
| ProblemType | probType |
| Problem type. | |
Definition at line 22 of file gprwrapper.h.
| gurls::GPRWrapper< T >::GPRWrapper | ( | const std::string & | name | ) |
| name | Name of the option's structure that will be initialized |
Definition at line 14 of file gprwrapper.hpp.
: KernelWrapper<T>(name), norm(NULL) { }
| gMat2D< T > * gurls::GPRWrapper< T >::eval | ( | const gMat2D< T > & | X | ) | [virtual] |
| [in] | X | Input matrix |
Implements gurls::GurlsWrapper< T >.
Definition at line 123 of file gprwrapper.hpp.
{
gMat2D<T> vars;
return eval(X, vars);
}
| gMat2D< T > * gurls::GPRWrapper< T >::eval | ( | const gMat2D< T > & | X, |
| gMat2D< T > & | vars | ||
| ) |
| [in] | X | Input matrix |
Definition at line 131 of file gprwrapper.hpp.
{
gMat2D<T> empty;
NormTestZScore<T> normTask;
PredGPRegr<T> predTask;
GurlsOptionsList *normX = normTask.execute(X, empty, *(norm));
gMat2D<T> &Xresc = normX->getOptValue<OptMatrix<gMat2D<T> > >("X");
PredKernelTrainTest<T> predkTrainTest;
this->opt->removeOpt("predkernel");
this->opt->addOpt("predkernel", predkTrainTest.execute(Xresc, empty, *(this->opt)));
GurlsOptionsList *pred = predTask.execute(Xresc, empty, *(this->opt));
delete normX;
OptMatrix<gMat2D<T> >* pmeans = pred->getOptAs<OptMatrix<gMat2D<T> > >("means");
pmeans->detachValue();
gMat2D<T> &predMeans = pmeans->getValue();
gMat2D<T> &predVars = pred->getOptValue<OptMatrix<gMat2D<T> > >("vars");
const unsigned long n = predMeans.rows();
const unsigned long t = predMeans.cols();
T* column = predMeans.getData();
const T* std_it = norm->getOptValue<OptMatrix<gMat2D<T> > >("stdY").getData();
const T* mean_it = norm->getOptValue<OptMatrix<gMat2D<T> > >("meanY").getData();
const T* pvars_it = predVars.getData();
vars.resize(n, t);
T* vars_it = vars.getData();
for(unsigned long i=0; i<t; ++i, column+=n, ++std_it, ++mean_it, vars_it+=n)
{
scal(n, *std_it, column, 1);
axpy(n, (T)1.0, mean_it, 0, column, 1);
copy(vars_it, pvars_it, n);
scal(n, (*std_it)*(*std_it), vars_it, 1);
}
delete pred;
return &predMeans;
}
| T gurls::GurlsWrapper< T >::eval | ( | const gVec< T > & | X, |
| unsigned long * | index = NULL |
||
| ) | [virtual, inherited] |
| [in] | X | Input point |
| [out] | index | Index of the estimated label |
Definition at line 26 of file wrapper.hpp.
{
if(!trainedModel())
throw gException("Error, Train Model First");
gMat2D<T>X_mat(1, X.getSize());
copy(X_mat.getData(), X.getData(), X.getSize());
gMat2D<T>* pred_mat = eval(X_mat);
const T* pred = pred_mat->getData();
const unsigned long size = pred_mat->getSize();
const T* max = std::max_element(pred, pred+size);
T ret = *max;
if(index != NULL)
*index = max-pred;
delete pred_mat;
return ret;
}
| void gurls::GurlsWrapper< T >::loadModel | ( | const std::string & | fileName | ) | [virtual, inherited] |
| fileName | name of the file containing the data to load |
Definition at line 61 of file wrapper.hpp.
| void gurls::GurlsWrapper< T >::saveModel | ( | const std::string & | fileName | ) | [virtual, inherited] |
| fileName | name of the file where data will be saved |
Definition at line 55 of file wrapper.hpp.
| void gurls::KernelWrapper< T >::setKernelType | ( | typename KernelWrapper< T >::KernelType | value | ) | [virtual, inherited] |
| value |
Definition at line 129 of file wrapper.hpp.
{
kType = value;
// std::string &type = this->opt->template getOptValue<OptString>("kernel.type");
// switch(value)
// {
// case RBF:
// type = std::string("rbf");
// case LINEAR:
// type = std::string("linear");
// case CHISQUARED:
// type = std::string("chisquared");
// }
}
| void gurls::GurlsWrapper< T >::setNparams | ( | unsigned long | value | ) | [virtual, inherited] |
| [in] | value |
Definition at line 67 of file wrapper.hpp.
| void gurls::KernelWrapper< T >::setNSigma | ( | unsigned long | value | ) | [virtual, inherited] |
| value |
Definition at line 161 of file wrapper.hpp.
| void gurls::GurlsWrapper< T >::setParam | ( | double | value | ) | [virtual, inherited] |
| [in] | value |
Reimplemented in gurls::NystromWrapper< T >.
Definition at line 79 of file wrapper.hpp.
{
if(!opt->hasOpt("paramsel"))
opt->addOpt("paramsel", new GurlsOptionsList("paramsel"));
if(opt->hasOpt("paramsel.lambdas"))
opt->getOptValue<OptMatrix<gMat2D<T> > >("paramsel.lambdas").getData()[0] = (T)value;
else
{
gMat2D<T> * lambdas = new gMat2D<T>(1,1);
lambdas->getData()[0] = (T)value;
opt->getOptAs<GurlsOptionsList>("paramsel")->addOpt("lambdas", new OptMatrix<gMat2D<T> >(*lambdas));
}
setNparams(1);
}
| void gurls::GurlsWrapper< T >::setProblemType | ( | typename GurlsWrapper< T >::ProblemType | value | ) | [virtual, inherited] |
| [in] | value |
Definition at line 103 of file wrapper.hpp.
{
probType = value;
opt->getOptValue<OptString>("hoperf") = (value == CLASSIFICATION)? "macroavg": "rmse";
}
| void gurls::KernelWrapper< T >::setSigma | ( | double | value | ) | [virtual, inherited] |
| value |
Definition at line 147 of file wrapper.hpp.
| void gurls::GurlsWrapper< T >::setSplitProportion | ( | double | value | ) | [virtual, inherited] |
| [in] | value |
Definition at line 97 of file wrapper.hpp.
{
opt->getOptValue<OptNumber>("hoproportion") = value;
}
| void gurls::GPRWrapper< T >::train | ( | const gMat2D< T > & | X, |
| const gMat2D< T > & | y | ||
| ) | [virtual] |
| X | Input data matrix |
| Y | Labels matrix |
Implements gurls::KernelWrapper< T >.
Definition at line 24 of file gprwrapper.hpp.
{
this->opt->removeOpt("split");
this->opt->removeOpt("optimizer");
if(norm != NULL)
{
delete norm;
norm = NULL;
}
NormZScore<T> taskNorm;
norm = taskNorm.execute(X, y, *(this->opt));
const unsigned long nlambda = static_cast<unsigned long>(this->opt->getOptAsNumber("nlambda"));
const unsigned long nsigma = static_cast<unsigned long>(this->opt->getOptAsNumber("nsigma"));
OptTaskSequence *seq = new OptTaskSequence();
GurlsOptionsList * process = new GurlsOptionsList("processes", false);
OptProcess* process1 = new OptProcess();
process->addOpt("one", process1);
this->opt->addOpt("seq", seq);
this->opt->addOpt("processes", process);
// this->opt->printAll();
if(this->kType == KernelWrapper<T>::LINEAR)
{
if(nlambda > 1ul)
{
*seq << "split:ho" << "kernel:linear" << "paramsel:hogpregr";
*process1 << GURLS::computeNsave << GURLS::computeNsave << GURLS::computeNsave;
}
else if(nlambda == 1ul)
{
if(this->opt->hasOpt("paramsel.lambdas"))
{
*seq << "kernel:linear";
*process1 << GURLS::computeNsave;
}
else
throw gException("Please set a valid value for the regularization parameter, calling setParam(value)");
}
else
throw gException("Please set a valid value for NParam, calling setNParam(value)");
}
else if(this->kType == KernelWrapper<T>::RBF)
{
if(nlambda > 1ul)
{
if(nsigma > 1ul)
{
*seq << "split:ho" << "paramsel:siglamhogpregr" << "kernel:rbf";
*process1 << GURLS::computeNsave << GURLS::computeNsave << GURLS::computeNsave;
}
else if(nsigma == 1ul)
{
if(this->opt->hasOpt("paramsel.sigma"))
{
*seq << "split:ho" << "kernel:rbf" << "paramsel:hogpregr";
*process1 << GURLS::computeNsave << GURLS::computeNsave << GURLS::computeNsave;
}
else
throw gException("Please set a valid value for the kernel parameter, calling setSigma(value)");
}
else
throw gException("Please set a valid value for NSigma, calling setNSigma(value)");
}
else if(nlambda == 1ul)
{
if(nsigma == 1ul)
{
if(this->opt->hasOpt("paramsel.sigma") && this->opt->hasOpt("paramsel.lambdas"))
{
*seq << "kernel:rbf";
*process1 << GURLS::computeNsave;
}
else
throw gException("Please set a valid value for kernel and regularization parameters, calling setParam(value) and setSigma(value)");
}
else
throw gException("Please set a valid value for NSigma, calling setNSigma(value)");
}
else
throw gException("Please set a valid value for NParam, calling setNParam(value)");
}
*seq << "optimizer:rlsgpregr";
*process1 << GURLS::computeNsave;
GURLS G;
G.run(norm->getOptValue<OptMatrix<gMat2D<T> > >("X"), norm->getOptValue<OptMatrix<gMat2D<T> > >("Y"), *(this->opt), "one");
}
| bool gurls::GurlsWrapper< T >::trainedModel | ( | ) | [protected, virtual, inherited] |
| value | |
| value |
Definition at line 111 of file wrapper.hpp.